Discriminating European cyprinid specimens by barcode high-resolution melting analysis (Bar-HRM)
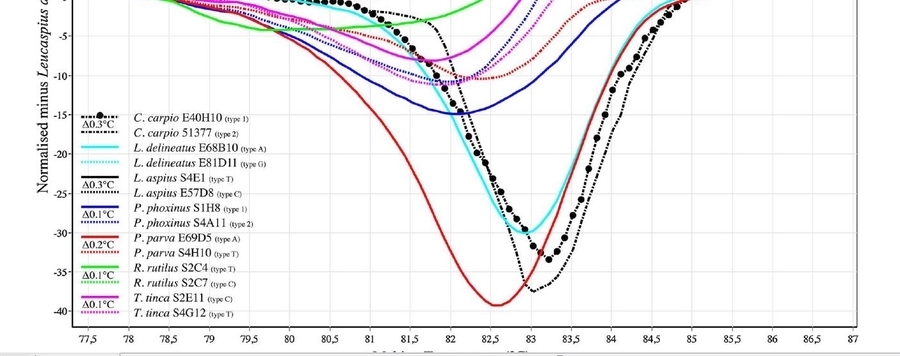
Today, one of the challenges facing modern biology is to develop an accurate, reliable and cost- efficient method for the rapid identification of organisms to species level. Here, we combine high resolution melting (HRM) analysis with DNA barcoding (Bar-HRM) and evaluated its efficiency to discriminate between 215 specimens of 22 common European cyprinid species. After screening the standard barcode region of the mitochondrial cytochrome oxidase gene subunit I (COI) for a distinct fragment, a short region (142 bp) with sufficient sequence diversity was chosen. Using the selected mini-barcode, Bar-HRM analysis was performed to test the potential of this closed- tube method to reveal genetic variation between the cyprinids. To evaluate inter-run reproducibility several Bar-HRM runs were performed on different days using between 8 and 10 specimens per species, resulting in only minor variations in the amplification curves and melting temperatures. The technique was also found to be highly sensitive to single base changes, allowing the detection of geographic variants. 92% of the specimens were unambiguously assigned to the correct cyprinid species whereas in seven individuals the presence of ambiguities prevents the assignment; in case of Barbus barbus the method failed. We provide a guideline for setting up a simple molecular identification tool for unknown samples from a pool of target species with one pair of mini-barcode primers and reduced contamination risk. We demonstrate that Bar-HRM may provide a reliable and simple alternative to Sanger-sequencing based identification in certain applications with the potential to significantly reduce time (up to 241 h) and labor costs (up to 88%) during the process of identification.






