Quick facts
Project title:
APPETITE - (A) (P)hylogenetic (P)ipeline (E)nabling (T)remendous (I)nvestigativ (T)ree (E)valuations
Description
APPETITE is a new automated process-pipeline for molecular phylogenetic purposes with nearly endless analytical possibilities. The pipeline is a modularly constructed system to the extent that each operational process is programmed as a single module.
Based on their main process characteristic within APPETITE, implemented modules are either defined as main (analysis) or help modules.
Main modules execute actual analysis steps, applying different software scripts and models. This includes, for example, processes of data simulation, multiple sequence alignments, alignment evaluation, tree reconstruction, and tree evaluation. Individual analysis steps are thereby further divided thematically into 'mainstream' modules (handling of external developed, widely distributed software) and 'non-mainstream' or 'alternative' modules (less widely distributed or new external approaches as well as new internal algorithmic developments).
Help modules are background tools in the pipeline, which organise a smooth operation flow between nested and combined main modules, like input/output file handling, table summaries, or graphical output, depending on the main module specific characteristics.
The third group of APPETITE modules encompasses core modules. Core modules are control units within APPETITE, each specifying an individually combined main module process structure with individually defined parameter settings for each main module. Core modules and parameter settings can be easily saved as text file and thus modified for new process chains or further proceeding subanalyses.
Following figures give a short introduction of the APPETITE modularly structure and example schemes about a simple and a more complex core module process chain, which are both only the tip of the iceberg of nearly unlimited possibilities of automized process chains within APPETITE.
Figure 1: Overview of the modular construction of APPETITE. Modules are divided thematically into main (analysis), help, and core modules. Each core module defines and controls a specific process chain of nested main module analysis steps, whereas main modules control specific analysis software scripts for phylogenetic purposes, e.g. alignment masking with AliSCORE. Help modules operate in the background, organizing a smooth operation flow between nested main module operations and provide enhanced output summaries of each process step. Core modules and parameter setups can be separately saved for reutilisation. The mentioned modules are only exemplary and do not correspond to all included methods.
Figure 2: Example of a simple process chain of main modules as implemented in the core module 'Munich', comprising MAFFT alignment, AliSCORE masking and AliCUT editing of single gene raw data, followed by FASconCAT-G concatenation and a Maximum Likelihood tree reconstruction of the FASconCAT-G generated multiple gene supermatrix. Individual output results and info of each process step are printed to main module assigned output folders.
Figure 3: Help module structure of the process chains defined by the core module 'Munich', including usage of multiple processing, in-/output conversion, and summary of single results.
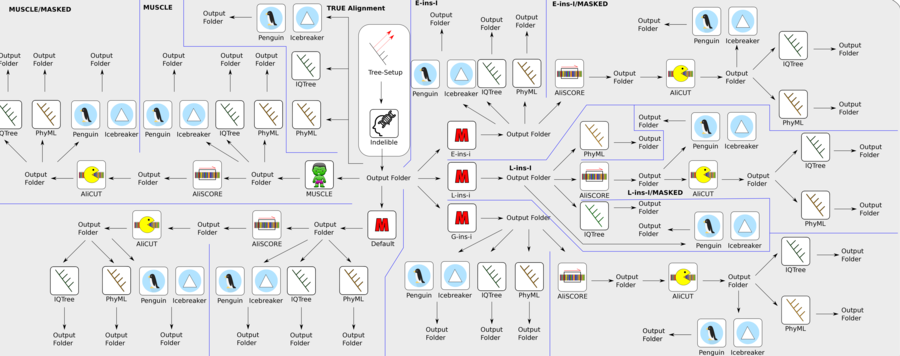
Figure 4: Example of a more complex core module process structure (core module 'Manchester'). Based on a set of input gene raw data, the core module operates different alignment strategies, followed by alignment quality assessments and further improvements. Finally, different tree reconstruction approaches are conducted for each of the different analysis strategies. All results and graphical outputs are summarised in different output folders and saved in a final pdf result document.
Overview of currently included main modules:
-
Data simulation
-
Sequence alignment methods
-
included: MAFFT, Muscle, T-Coffee, Clustal Omega, ProbCons, Dialign-TX, POA (Partial Order Alignment)
-
Tree reconstruction methods
-
included: PhyML, IQ-Tree, IQPNNI, PAUP, Penguin
-
Data evaluation methods
-
included: HoT, AliScore, Aligroove, Penguin, MARE, BACoCA, CompareTrees(CoNe), JOIN, T-Coffee tree
Location