DiStats
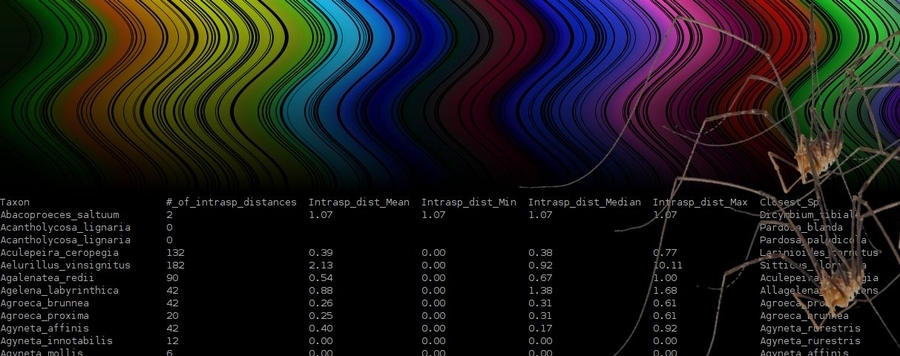
The Perl script DiStats simplifies the task of preparing DNA barcode data releases. Among other statistics, it computes intraspecific distances, data on closest species pairs, or on most distant congeners. DiStats expects a FASTA input, calculates p-distances or K2P and can be parallelized. It can produce two output files: a table with statistics for each species plus optionally also the matrix of all pairwise distances in the dataset. For an alignment containing 1000 COI barcode sequences, the analysis will take around 6 minutes when using a single thread (on a 3.4 GHz processor). DiStats has an algorithmic complexity (O) of approximately O(n^2), which means that run time increases exponentially with the number of input sequences (n). Using multiple CPU threads reduces the run time by a factor of 1/c, where c is the number of threads.
Documentation included.
Cite as:
Astrin, Höfer, Spelda, Holstein, Bayer, Hendrich, Huber, Kielhorn, Krammer, Lemke, Monje, Morinière, Rulik, Petersen, Janssen, Muster (2016). Towards a DNA barcode reference database for spiders and harvestmen of Germany. PLoS ONE 11(9): e0162624.
doi:10.1371/journal.pone.0162624
Programming by:
Hannah Janssen & Malte Petersen






