AliGROOVE
AliGROOVE determines and visualizes the content of potentially randomized sequence similarities and alignment ambiguities in multiple sequence alignments.
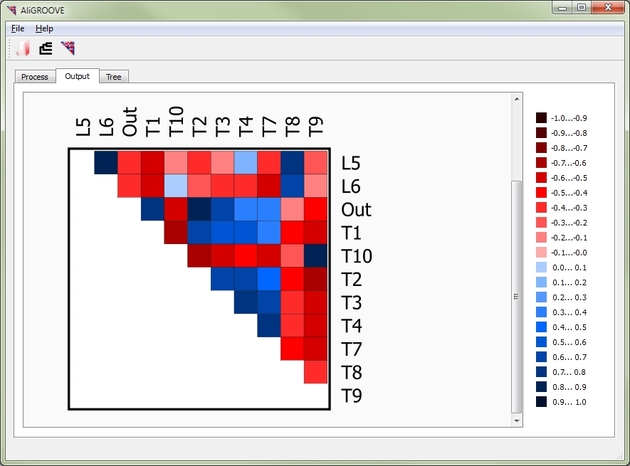
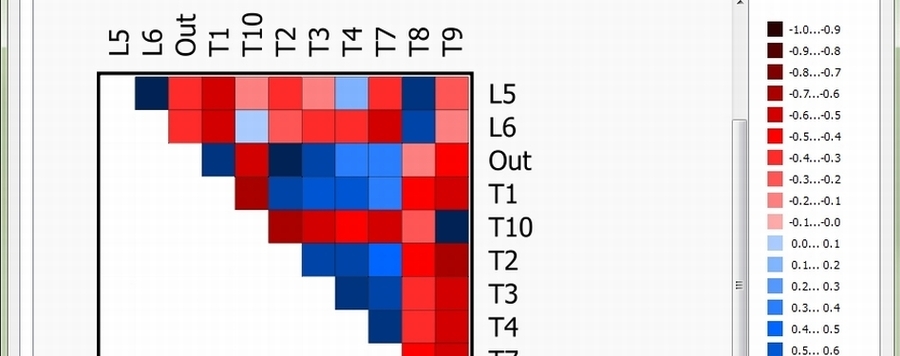
The AliGROOVE algorithm is an adaptation of the recently published ALISCORE masking algorithm (Misof & Misof, 2009, Kück et al., 2010). AliGROOVE summarizes site scores of profiles of sequence similarity normalized over the whole alignment length from each pairwise comparison and translates the obtained scoring distances between sequences into a similarity Matrix
(Figure 1).
AliGROOVE can be used to analyse sequence heterogeneity of concatenated supermatrices, but also to analyse the influence of heterogeneity that derives from single gene partitions. Therefore, the script is well suited for single gene analyses as well as for phylogenomic data.
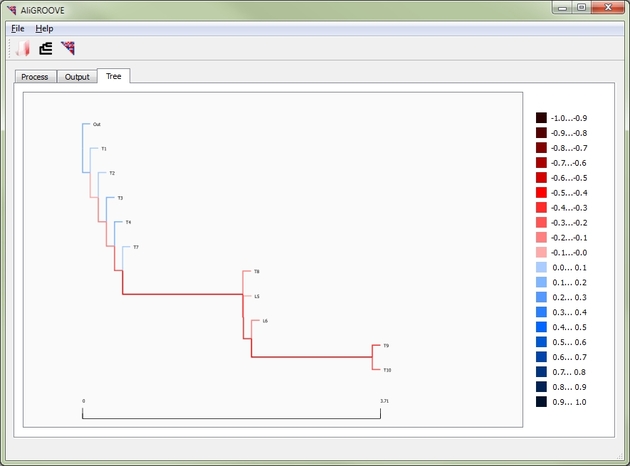
In addition, AliGROOVE can tag unreliable branches of given topologies. To define the reliability of single branches, AliGROOVE calculates the average similarity between taxa which are connected by a respective branch. This tagging of branches is an indirect estimation of reliability of a subset of all possible splits guided by a topology. Calculated reliabilities of single branches are shown colorized in tree Output (Figure 2).